
CryoEM Structure Refinement by Integrating NMR Chemical Shifts with Molecular Dynamics Simulations

Protocol for the structure determination of a CA hexamer using MAS NMR
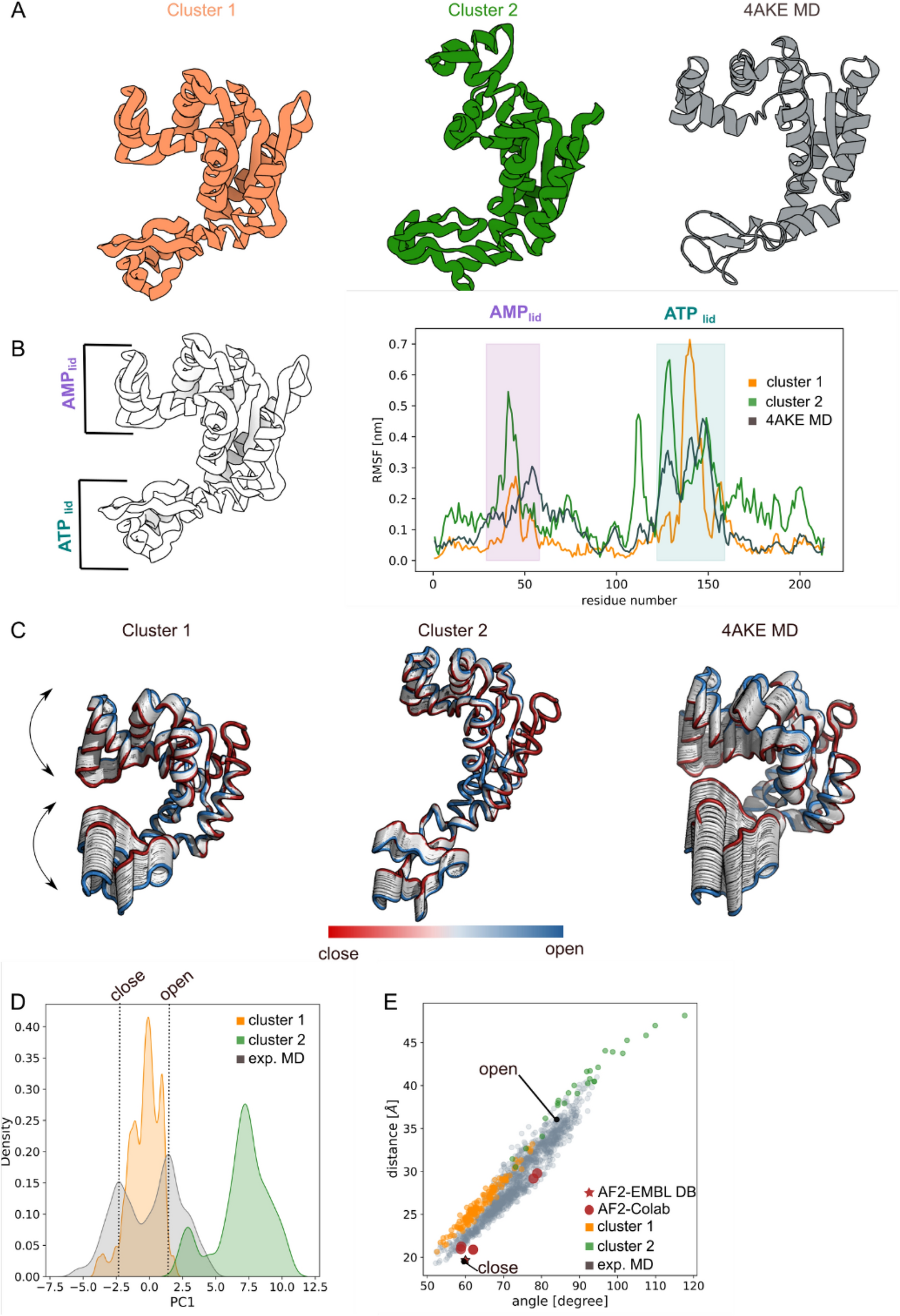
Machine learning/molecular dynamic protein structure prediction approach to investigate the protein conformational ensemble

Integrating experimental data with molecular simulations to investigate RNA structural dynamics - ScienceDirect

CryoEM Structure Refinement by Integrating NMR Chemical Shifts with Molecular Dynamics Simulations

Imaging active site chemistry and protonation states: NMR crystallography of the tryptophan synthase α-aminoacrylate intermediate

Integrated NMR/EM structure determination approach. a In step 1, 40

Structure refinement of ribosome-binding element from turnip crinkle

Chemical shift-based methods in NMR structure determination - ScienceDirect

Experimental NMR and EM data of the 468 kDa TET2 assembly. a MAS NMR

CryoEM Structure Refinement by Integrating NMR Chemical Shifts with Molecular Dynamics Simulations

PDB 5upw structure summary ‹ Protein Data Bank in Europe (PDBe) ‹ EMBL-EBI

Using NMR Chemical Shifts and Cryo-EM Density Restraints in Iterative Rosetta-MD Protein Structure Refinement

Computational biophysics meets cryo‐EM revolution in the search for the functional dynamics of biomolecular systems - Costa - 2024 - WIREs Computational Molecular Science - Wiley Online Library
Two CRPL Research Poster Submissions Accepted for SC18








